Received: Thu 07, Sep 2023
Accepted: Tue 26, Sep 2023
Abstract
The causes of the alterations found in the brains of patients with alzheimer's disease (AD) begin before the first signs of memory loss appear, and are still unclear. Adequate research models are essential to understand the mechanisms that cause the onset of these alterations, as well as to advance in the diagnosis, development and testing of treatments for the AD. Animal research models fail to recreate the great diversity and complexity inherent to the human brain, so in vitro systems based on human pluripotent stem cells (hPSCs) present themselves as an important alternative. Differentiation of hPSCs into two-dimensional (2D) cell culture models allows recreation of various brain functional processes and the three-dimensional (3D) cell culture models or human brain organoids (hCOs) recapitulate the cellular diversity and structure of the human brain. hCOs from human induced pluripotent stem cells (hiPSCs) from patients with familial (APP, PSEN1 and PSEN2 mutations) or sporadic AD allow identifying and studying changes due to this pathology. This review presents an overview of the research models used to study the AD, and recapitulates the advantages and discusses the challenges of the hCOs as an innovative and promising technology that will aid in the understanding of AD.
Keywords
Alzheimer's disease, human cerebral organoids, hiPSCs, pluripotent stem cells
1. Introduction
Alzheimer's disease (AD) is the principal cause of dementia in the elderly population and one of the leading causes of death worldwide [1-4]. The AD incidence is expected to increase dramatically by 2050 mainly due to the rise in life expectancy of the world population [5, 6]. Moreover, AD is a disabling disorder, and the maintenance of the patients reaches a high cost for families and society, which makes AD a problem for public health [6, 7]. In the last decades, great advances have been made in the field of AD, but the main causes that trigger AD and its etiology are still unclear.
The neurodegenerative process of AD appears in adults and older people, beginning with some progressive memory impairments that involve alteration of cognitive processes, self-awareness and memory loss. In later stages, AD patients also manifest aphasia, amnesia, agnosia and apraxia [8-11]. The neurological changes are accompanied of two main histopathological features observed in the post-mortem brains of AD patients: the abnormal intracellular hyperphosphorylation of Tau protein (p-Tau) and the presence of extracellular amyloid plaques formed by accumulation of the long variants of β-amyloid (Aβ) peptide. On the one hand, p-Tau protein would lead to the formation of aggregates or neurofibrillary tangles (NFTs) inside the cells that would cause failures in the normal functioning of the neurons. By the other hand, Aβ peptide first would appear extracellularly as monomers that would associate with each other, forming oligomers and finally their aggregation would give rise to senile plaques (Figure 1) [8, 11-13]. Recent studies indicate an interrelation between both alterations found in AD [14, 15].
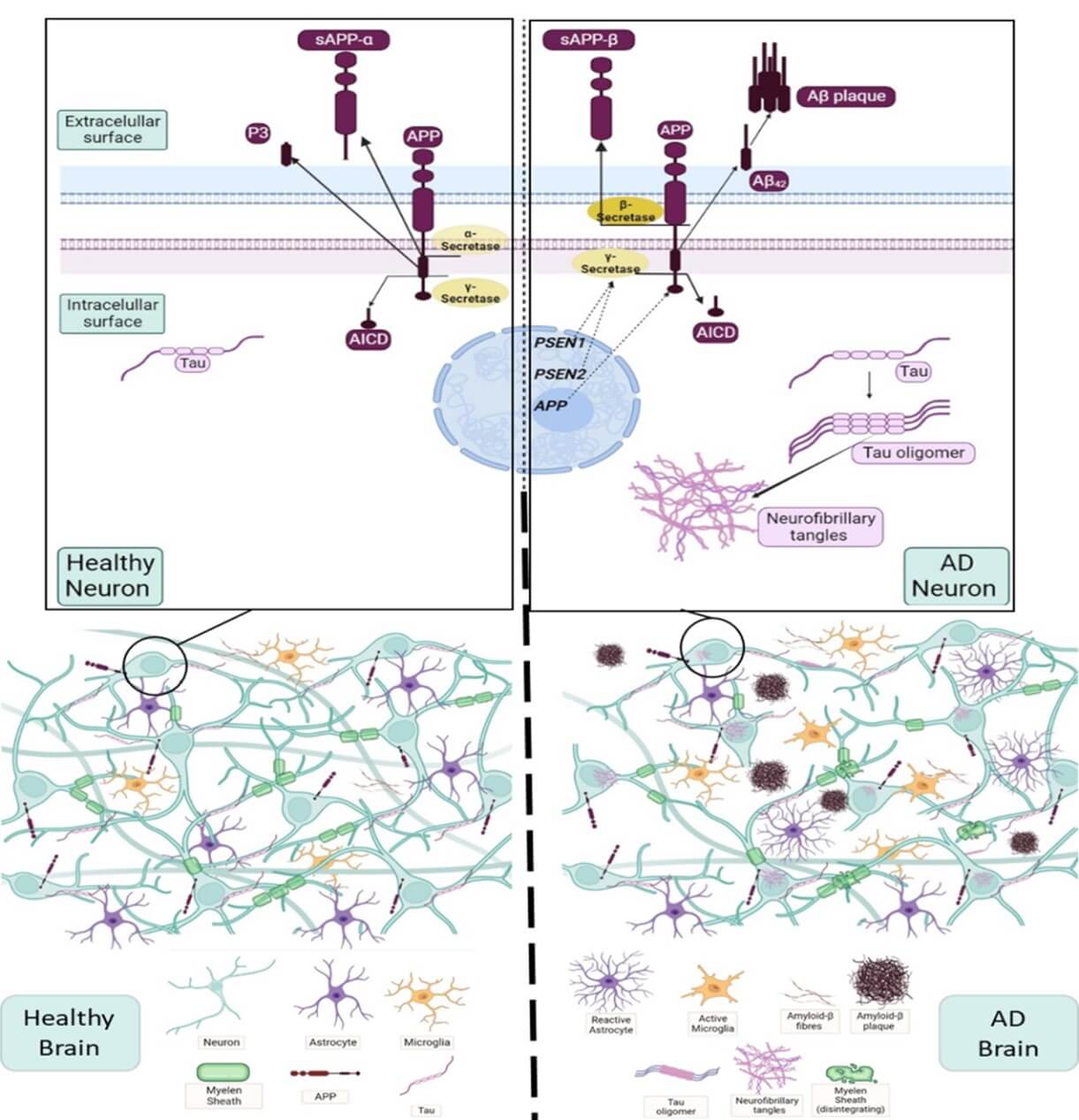
In the brain with AD (right) in comparison to a normal brain (left) the main physiological changes found are: presence of Aβ fibers and plaques, NFTs of p-Tau, damage of the myelin sheath, disintegration and reduction of dendrites in neurons, activation of microglia and presence of reactive astrocytes. The presence of Aβ plaques and NFTs in the neuron with AD is shown in the upper right of the figure together with the schema of amyloidogenic pathway of amyloid precursor protein (APP) (β-secretase pathway) and the genes that encode the catalytic subunit of γ-secretase, PSEN1 (presenilin-1) and PSEN2 (presenilin-2) implicated in the development of familial AD.
Both events (NFTs of p-Tau and senile plaques of Aβ peptide) appear in patients with sporadic AD (sAD) and familial AD (fAD). At least 90% of all cases of AD are sporadic and occurs in patients with more than 65 years. It is associated with aging, the allelic composition of apolipoprotein E (APOE), failures in the correct functioning of microglia and brain metabolism [16-19]. The prevalence of fAD is less than 10% of the AD cases and appears in patients with ages younger than 65 years. It is due to autosomal dominant mutations on the gen of the amyloid precursor protein (APP, located on chromosome 21) and mutations on genes of Presenilins 1 and 2 (PSEN1 and PSEN2, located on chromosome 14 and chromosome 1, respectively). It should be noted that APP is the precursor protein of Aβ peptides (Aβ40 and Aβ42) and Presenilins 1 and 2 are part of the γ-secretase catalytic complex, one of the enzymes that cleaves APP during its proteolytic processing. It is important to highlight that most of the mutations described for APP, PSEN1 and PSEN2 genes favor production and accumulation of Aβ peptide in the brain parenchyma of AD patients [11, 20].
Probably because the pathogenic mechanisms that cause AD have not yet been determined currently, there is no effective cure for this disease. In recent decades, only a few drugs have been approved for the palliative treatment of AD, such as inhibitors of acetylcholinesterase (donepezil, galantamine or rivastigmine) or antagonists of N-methyl-D-aspartate receptor (memantine) [21-23]. More recently, the FDA has approved a controversial new treatment, aducanumab (Aduhelm™), an anti-Aβ peptide monoclonal antibody, hoping to reduce their accumulation in brains of AD patients [24, 25]. However, none of these pharmacological treatments is capable of stopping the neurodegenerative process in AD.
Because the causes of this disease are still not well understood, many efforts are being carried out to know the origin, development and evolution of AD. One of the reasons why the causes of the disease are still not well known is the absence of good study models that allow replicating this disease. Animal models with human mutations (in APP, PSEN1 or PSEN2 genes) have been used to approach the study of AD. Nevertheless, there are many differences regarding the development of the disease in humans [26-29]. Secondly, the technology of human stem cells has allowed progress in the knowledge of AD, especially the use of induced pluripotent stem cells (iPSCs) derived from AD patients [10, 30-32]. However, the two-dimensional (2D) models have limitations, such as the lack of three-dimensional (3D) organization and cell diversity that make up the human brain [33, 34]. For this reason, the development and use of human cerebral organoids (hCOs) can be a good alternative to deepen and advance in the pathogenic mechanisms of AD.
In this review, we present the milestones reached in relation to the study of AD with animal models and in vitro models. Finally, we will focus on the use of hCOs as a promising technology to study and model of both sAD and fAD.
2. Animal Models of Alzheimer’s Disease (AD)
Several animal species (Drosophila melanogaster, Caenorhabditis elegans, Danio rerio), but mainly mice, have been used during last decades to understand the pathology of AD [29]. Most of the studies have focused on APP mutations [27-29], although mutations in other genes have also been analyzed, like PSEN1, APOE or MAPT [29]. For studies at behavioral, phenotypical and morphological level, these models have been very useful; however, they have shown limitations in modelling AD. In fact, the failure rate with these models in drug discovery has been of 99% [26, 35]. In addition, they are not realistic enough because neurodegeneration, like neuronal loss or the development of NFTs, cannot be observed in their brains.
Furthermore, there are important anatomical differences between brain morphology [29, 36-38] and physiology of the mouse and human brain, as the organization of ventricular zones. Animal models have a bias in the information they provide, since the genes that are modified only represent a minority of cases (fAD) and do not represent the totality of the pathology [22].
3. In vitro Models of Alzheimer Disease (AD)
3.1. Stem Cells and Adherent Models (2D)
Recent advances in the field of stem cells and new knowledge related to cell reprogramming have opened up a new world of opportunities and the creation of more reliable models for a better understanding of the etiological and molecular process of AD. During the last decades, the study and characteristics of stem cells has been deepened, as well as their possible therapeutic applications or as an alternative method to in vivo procedures [39, 40].
Stem cells are defined as cells with a high renewal potential and the capacity to differentiate into more specialized cell types [41]. There are four types of stem cells depending on how many different types of cells can be obtained from them: totipotent, pluripotent, multipotent and unipotent [42]. The most used in research are the multipotent (MSCs) and pluripotent stem cells (PSCs) [23, 37, 41]. MSCs can be derived from foetal, neonatal or adult tissues, but also from PSCs [42]. PSCs can be obtained from embryonic stem cells (ESCs) or by reprogramming somatic cells to induced pluripotent stem cells (iPSCs) [41].
The technique for creating iPSCs from fibroblasts was developed by Takahashi and Yamanaka, through a retroviral infection, that allowed the introduction of the factors Oct4, Sox2, Klf4 and c-Myc [40]. The same year, the group of Yu et al. also reported the reprogramming of human somatic cells to PSCs by the use of the Oct4, Nanog, Sox2 and Lin28 factors [43]. In recent years, much progress has been made in reprogramming techniques, to the point of obtaining safer and more efficient non-integrative methods [44-46].
In 2011 were designed and used in research the first iPSCs derived from patients with fAD [47, 48], and a year later from patients with sAD [30]. In these studies the presence of PSEN1 mutations found in fAD patients were associated with accumulation of Aβ peptides due to alterations in the ubiquitin kinase system [49], γ-secretase activity [50], Tau proteostasis [51] and up-regulation of calcium-controlling receptors in the endoplasmic reticulum (ER) [52]. In the early stages of AD were also observed diverse alterations in astrocytes [53-55].
Studies with iPSCs from fAD patients with APP mutations found that some iPSCs are more prone than others are in accumulation and aggregation of Aβ peptides [56]. APP alterations were studied in relation to p-Tau levels [57], cholesterol receptors involved in endocytosis and clearance of Aβ [58], and mitochondrial dysregulation [59]. Because of these studies, new treatments have been proposed to reduce Aβ peptides such as the use of statins [60], or various chemicals to increase Tau autophagy [61]. Despite these studies, many of the functions of APP involved in the disease are still unknown [62]. Another approach to study AD is to use iPSCs from individuals with down syndrome (DS), who present in most cases with early-onset dementia similar to patients with fAD due to the triplication of chromosome 21, where APP is encoded [63-65]. Deletion in these models of the extra copy of APP reverses and normalizes the Aβ40/Aβ42 ratio [66].
iPSCs from sAD patients has allowed the study of the APOE gene increasing the ratio of Aβ42 similar to what was observed in post-mortem brains [67, 68]. Not all APOE gene isoforms are aggressive; the APOE2 is neuroprotective, whereas APOE4 is the most toxic [69]. Several studies have found APOE4 to be associated with proinflammatory profiles and increased TREM2 [70], and hPSC-derived astrocytes with APOE mutations are generated for drug screening [71]. Recently a link between the APOE4 and the SARS-CoV-2 infection has been established with increased neurodegeneration and synapse loss [72].
Other 14 genes with direct involvement in fAD have been described with these models [73]. In addition to the alterations described above, increased oxidative damage [74] leading to alterations in membrane permeability [75] has been found. Microglia is also affected, due to failures in TREM2 [76], with an increase of microglial cells involved in the clearance of Aβ and Tau around Aβ accumulations in AD patients [77, 78]. For phagocytosis to be successful, support from mitochondria is required to mobilise all the proteins involved [79], although these also end up being affected by increased reciprocal Aβ42 and Tau deposition [80]. The degradation of Aβ, likewise, is mediated by insulin [81], so its accumulation induces insulin resistance, along with toxic and apoptotic reactions, increasing AD symptoms [82]. Oher studies focus in the regulation of miRNAs that could protect against AD symptoms, such as the miRNA124 [83].
2D stem cell models have been a great advance in the study of neurodegenerative diseases. However, in relation to AD, there is a need to recreate the different stages in time and space, as well as some key events such as the aggregation of extracellular peptides. The appearance of the 3D models, which are now possible due to advances in stem cell technology and knowledge of iPSCs, are closer to simulate that occur in vivo and open the research for understanding AD [33, 34].
3.2. 3D Models: Human Cerebral Organoids
In addition to 2D cultures, the ability to develop hCOs revolutionized the field and reduced the use of other techniques. Lancaster and collaborators [84] made the first hCO protocol, with the main goal of understand the development of the human brain. This first hCO protocol from human PSCs could recreate different brain regions capable of influencing each other. The protocols in which organoids generate various brain regions spontaneously are called unguided protocols [84-86]. The authors defined organoids as 3D cultures with two or more cell types in an order and function similar to those that would be found in the organ [85]. Another simple definition can be a primitive organ that is obtained from a tissue sample using PSCs [87].
Currently, other type of protocols for the generation of hCOs exists. Guided protocols employ patterning factors that give rise to organoids of a specific brain region [88-95]. For example, cortical organoids have the possibility of combining them to achieve a better understanding of cell, synapse, and neuron interaction and astrocyte maturation in these complex structures. In addition, this type of protocols can be used without the need of support [89, 90]. Table 1 lists the main publications with hCOs to study AD. Moreover, the identity of hCOs with the human foetal cerebral cortex has been confirmed [96]. Other studies indicate that 45-day-old organoids have a large number of pathways and functions typical of neural tissue: metabolism, cell-cell adhesion, development of the cerebral cortex, organisation of the cytoskeleton, and others like the human brain at 16 weeks gestational age [32].
TABLE 1: Summary of the main publications with hCOs as research models to study
AD, classified attending to the protocol employed to generate the hCOs
(non-guided or guided) with mutations corresponding to familial AD (fAD),
sporadic AD (sAD), or other type.
|
Protocol |
aOrigin
human cell line |
bMutations |
cAD
Pathology |
dCell
types |
Studies |
Refs |
||
|
fAD non-guided hCOs |
||||||||
|
Lancaster, 2013 |
AD patient & DS Fibroblasts iPSCs |
PSEN1 (1) |
Aβ42/40 ratio Aβ Agg p-Tau Tau Agg |
Neurons Astrocytes |
In hCOs found
elevated levels of Aβ,
p-Tau and cell death similar to those observed in AD and DS brains |
[111] |
||
|
Lancaster, 2013
(STEMdiff™) |
AD patient Dermal Biopsy iPSCs |
APP (2) PSEN1 (3, 4) |
Aβ42/40 ratio |
Neurons |
Neuronal
hyperactivity due to increased VGLUT1 and decreased VGAT expression Nytrosynapain reduces neuronal hyperactivity |
[105, 108] |
||
|
Lancaster, 2013
(STEMdiff™) |
AD patient Fibroblasts iPSCs |
PSEN2 (5) |
Aβ42/40 ratio |
Neurons |
Smaller organoid size
and calcium dysregulation in AD hCOs versus control hCOs |
[103] |
||
|
Lancaster, 2013 |
AD patient Fibroblasts iPSCs |
APP (6) PSEN1 (7, 8, 9, 10, 11) |
Aβ38, Aβ40 Aβ42, Aβ43 |
Neurons |
PSEN1 and APP
mutations alter the neurogenesis and Aβ secretome, and favour aging and neurodegeneration |
[104, 106] |
||
|
fAD guided hCOs |
||||||||
|
Kadoshima, 2013 (modified)
|
AD patient Fibroblasts iPSCs |
APP dup PSEN1 (1, 7) |
Aβ42/40 ratio Aβ Agg p-Tau, Tau Alt Endosomes |
Neurons |
hCOs recapitulate the
AD pathology and the use of β-
and γ-secretase inhibitors
significantly reduces this pathology |
[97] |
||
|
Qian, 2016 |
AD patient Fibroblasts iPSCs |
APP (6) PSEN1 (8,9 ,10) |
Aβ Agg p-Tau |
Neurons |
A risk factor of fAD is the failure of the genetic regulator
5-hydroxymethylcytosine |
[110] |
||
|
Qian, 2016 (modified) |
AD patient Fibroblasts iPSCs |
PSEN1 (1) PSEN2 (5) |
Aβ Agg Tau Agg |
Neurons Astrocytes |
Using an adenovirus
to introduce Tau into fAD hCOs enhances aggregation and phosphorylation of
the Tau protein |
[102] |
||
|
Park, 2023 |
AD patient Fibroblasts iPSCs |
PSEN1 |
Aβ Agg Alt astrocytes Alt dendrites |
Neurons Astrocytes |
Spheroid protocol to
use hCOs in pharmacological assays |
[99] |
||
|
Yan, 2018 |
AD patient Fibroblasts iPSCs |
PSEN1 (4) |
Aβ42 p-Tau
Neuroinflammation |
Neurons |
hCOs to study of the
microenvironment in AD brains |
[100] |
||
|
Paşca, 2015 (modified) |
AD patient Fibroblasts |
APP (6)
PSEN1 (11) |
Aβ42/40 ratio p-Tau |
Neurons Astrocytes |
Hippocampal hCOs for
the study of AD. NeuroD1 to help in AD gene therapy |
[109] |
||
|
Sloan, 2018 |
AD patient Fibroblasts |
PSEN1 (12) |
Aβ Agg p-Tau |
Neurons |
Nanoparticles, as
STB-MP, help to reduce the pathology of the AD |
[101] |
||
|
Amin, 2018 Birey, 2017 |
AD patient Fibroblasts iPSCs |
PSEN1 (13, 14, 15) |
Aβ42/40 ratio p-Tau |
Neurons |
Mutations in PSEN1 increase the Notch pathway,
promoting generation of neuronal precursors |
[107] |
||
|
sAD non-guided hCOs |
|
|||||||
|
Lancaster, 2013 |
AD patient Skin Biopsy iPSCs |
PITRM1 KO |
Aβ42/40 ratio p-Tau |
Neurons |
Absence of PITRM1 causes a pathology similar to
AD due to mitochondria dysregulation |
[126] |
||
|
Lancaster, 2013
(STEMdiff™) |
AD patient Skin Biopsy iPSCs |
APOE ε3/ε3 APOE ε4/ε4 APOE KO |
Aβ Agg p-Tau |
Neurons Astrocytes |
APOE4 increases p-Tau as
well as cell apoptosis. APOE-/- hCOs to study pathological mechanisms in AD |
[19, 123] |
||
|
Lancaster, 2013
(STEMdiff™) |
AD patient PBMCs iPSCs |
APOE ε3/ε3 ε4/ε4 |
Aβ42/40 ratio p-Tau, Tau
|
Neurons Astrocytes |
Development a
platform to evaluated new drug to treat AD |
[118] |
||
|
Lancaster, 2013
(STEMdiff™) |
Healthy Fibroblasts iPSCs |
APOE ε3/ε3 APOE ε4/ε4 |
Aβ secretion p-Tau |
Neurons Astrocytes |
APOE4 neurons exhibited an
increase of the synapsis |
[121] |
||
|
Lancaster, 2013
(STEMdiff™) |
AD patient Fibroblasts iPSCs |
APOE ε3/ε3 APOE ε4/ε4 |
Aβ40 p-Tau |
Neurons Astrocytes |
APOE4 produces
neuronal dysregulation due to the
reduction of translocation to the nucleus of the repressor silencing
transcription factor (REST) |
[114] |
||
|
Lancaster, 2013
(STEMdiff™) |
Healthy Fibroblasts iPSCs |
APOE ε3/ε3
APOE ε4/ε4 |
Aβ Agg p-Tau Alt lipids |
Neurons Astrocytes |
Role of APOE4 expression in neurons and
astrocytes related to AD pathology |
[115] |
||
|
Lancaster, 2013 (modified) |
H9 hESCs |
ND |
Aβ Agg Neuroinflammation |
Neurons Astrocytes |
Herpes Simplex Virus
1HSV-1 induces AD like pathology |
[128] |
||
|
Lancaster, 2013
(STEMdiff™) |
Control ASE-9109 iPSCs |
BIN1 KO |
APP |
Neurons Astrocytes Oligodendrocytes |
BIN1 KO hCOs present smaller primary endosomes, and
early AD pathology |
[124] |
||
|
sAD guided hCOs |
|
|||||||
|
Raja, 2016 |
AD patient Fibroblasts iPSCs |
APOE ε3/ε3
APOE ε4/ε4 |
Aβ Agg p-Tau |
Neurons Astrocytes Microglia |
Transform of APOE4 hCOs to APOE3 hCOs reduces AD symptomatology |
[113] |
||
|
Paşca, 2015 |
AD patient PBMCs iPSCs |
NS |
Aβ Agg Tau Agg |
Neurons Astrocytes |
First neuro-spheroid model
derived from AD patients' blood |
[116] |
||
|
Paşca, 2015 |
AD patient PBMCs iPSCs |
NS |
Neuroinflammation |
Neurons |
Found of deregulated
proteins in AD related with axon development, platelet aggregation, RNA translation
and inflammation |
[117] |
||
|
Lin & Chen, 2008 Seki, 2012 Paşca, 2015 |
AD patient Fibroblasts iPSCs |
NS |
Aβ Agg p-Tau |
Neurons |
CKD-504 (HDAC6 selective
inhibitor) reduces levels of p-Tau |
[120] |
||
|
Other non-guided hCOs |
|
|||||||
|
Lancaster, 2013 |
DS Fibroblasts iPSCs |
APP dup |
Aβ Agg |
Neurons Astrocytes |
BACE2 as a dose-sensitive
AD-suppressor gene |
[139] |
||
|
Lancaster, 2013 Sheridan, 2012 |
Fibroblasts iPSCs (CRL-2522) |
ND |
Aβ Agg |
Neurons Astrocytes |
Treatment with Aftin-5 (β-42 inducer) modulates the posttranslational pathways of APP |
[132] |
||
|
Pavoni 2018 |
Fibroblasts iPSCs (CRL-2522) |
Tau (16) |
p-Tau |
Neurons Astrocytes |
Overexpression of Tau to
study neurodegenerative disease |
[137] |
||
|
Lancaster, 2013 |
Fibroblasts iPSCs |
ND |
Aβ Agg Tau Agg |
Neurons |
Treatment with serum
obtained from post-mortem AD patients increases Aβ and Tau in hCOs |
[135] |
||
|
Lancaster, 2013 (modified) |
PBMCs iPSCs |
ND |
Aβ Agg
p-Tau Alt Endosomes |
Neurons |
hCOs co-stimulated with Aβ. Estrogens reduce AD
symptomatology |
[133] |
||
|
Lancaster, 2013
(STEMdiff™) |
hESCs (UE02302) |
APP (2) BACE2 (17) |
Aβ42
Neural apoptosis |
Neurons Astrocytes |
The loss-function of BACE2 induce AD-like pathology |
[140] |
||
|
Other guided hCOs |
|
|||||||
|
Raja, 2016 |
Patient FTD iPSCs |
Tau (18) |
Tau Agg |
Neurons |
Inhibition of p25 reduces
p-Tau |
[136] |
||
|
Camp, 2015 Trujillo, 2019 Yao, 2020 |
DS iPSCs (DS1-iPS4) |
ND |
Aβ Agg Tau Agg |
Neurons |
DS hCOs show accumulation
of Aβ42 and Tau, as observed in AD |
[112] |
||
|
Zhang, 2023 |
iXCells Biotechnologies iPSCs |
ND |
Aβ42 Alt
Mitophagy |
NS |
hCOs treated with Aβ42. Galangin reduce Aβ42 mitophagy and
produce an increment of PTEN-induced kinase 1 (PINK1) |
[134] |
||
aiPSCs: induced Pluripotent Stem Cells; hESCs: human Embrionic Stem Cells;
DS: Down Syndrome; PBMCs: Peripheral Blood Mononuclear Cells; FTD:
Frontotemporal Dementia.
b1: A264E; 2: Swedish; 3: ΔE9; 4: M146V; 5: N141I;
6: London; 7: M146I; 8: int4del; 9: Y155H; 10: M139V; 11: R278K; 12: 14q24; 13:
L435F; 14: M146L; 15: D385A; 16: P301S; 17: G446R; 18: P301L; KO: Knock Out;
dup: duplication; ND: Not Disease mutation.
cAβ42/40:
Aβ42/ Aβ 40;
p-Tau: hyperphosphorylation of Tau protein; Agg:
Aggregates; Alt: Alterated.
dNS:
Not Specified.
3.2.1. Familial Alzheimer Disease (fAD)
One of the first AD studies performed using COs was that of the Raja group, using different hiPSCs from patients with fAD (APP duplication or PSEN1 mutations) and the protocol described by the Kadoshima group in 2013. In this work, AD-associated pathology was observed in these organoids, such as amyloid aggregation, p-Tau protein and endosome abnormalities compared to controls. They also observed that the use of β-secretase and γ-secretase inhibitors resulted in a reduction of Aβ and p-Tau [97].
Many authors have focused on studying diverse PSEN1 mutations that appears in most of the fAD cases [98]. Using guided hCOs models, researchers have observed AD pathology events as increased levels of Aβ42, decreased neural dendrite size [99], elevated gene expression of proinflammatory cytokines (IL-6 and TNF-α), upregulated syndecan-3, altered expression of matrix proteins and prominent levels of p-Tau [100]. In addition, some research models of hCOs of neurons and astrocytes from AD patients have also been proposed to test drug efficacy reducing Aβ aggregation [99]. hCOs has been used to study the microenvironment and test how to decrease the pro-inflammatory profile found in AD [100], and also as screening platform for novel AD treatment assessments as the effect of nanoparticles [101].
PSEN2 mutation has hardly been considered in hCOs, probably due to its lower incidence [98]. The few studies using PSEN2 mutations have observed an increase in Aβ42/Aβ40 ratio, increased p-Tau, asynchronous calcium transients, enhanced neuronal activity, and smaller hCOs size [102, 103]. An increase in caspase-3 in PSEN2 hCOs has also been detected associated with a high level of apoptosis [103]. The increase in the levels of p-Tau with adeno-associated virus was used in these PSEN2 hCOs as a research model to study tauopathies [102].
In addition to analysing the effect produced by mutations in PSEN1, it is interesting to compare them with the effects found due to mutations or duplication of APP, also present in fAD. hCOs with mutations in PSEN1 or in APP, generated with unguided protocol, show an increase in the Aβ42/ Aβ40 ratio, although present differences in the length of the Aβ fragments [104], and in the ratios [105]. hCOs with some mutations in PSEN1 produce premature neuronal differentiation, possibly due to a reduction in the notch pathway [106]. However, other PSEN1 mutations show upregulation of the notch pathway leading to an increase in neural progenitors as well as the reduction of post-mitotic neurons [107]. These PSEN1 mutations causing alterations in the notch pathway suggest that neural stem cell biology is affected in aging and disease. Regardless of the mutation, all fAD hCOs exhibit synaptic dysfunction, showing increased expression of VGLUT1 and decreased VGAT. In addition, further studies have observed that NitroSynapsin could be a good candidate to palliate these alterations [105, 108].
Other authors have opted to compare PSEN1 and APP mutations using hCOs from guided protocols. They observe Aβ aggregation, increase in the Aβ42/Aβ40 ratio, alterations in synaptic proteins or increase in p-Tau [109, 110], as seen using hCOs from non-guided protocols [104-106, 108]. Differences in the alterations are found also depending of the mutation [104, 109]. Alterations at the epigenetic level have been found using these models with a decrease in 5-hydroxymethylcytosine [110] and an increase in miRNA125b. The alterations at the mitochondrial level appear to be able to be regulated using NeuroD1 gene-based therapy [109].
Apart from using iPSCs from AD patients, it is also possible to use lines derived from down Syndrome (DS), since the extra copy of APP, producing as PSEN1 hCOs, presence of Aβ plaques, NTFs [111], or p-Tau [112], regardless of the protocol used to generate hCOs.
3.2.2. Sporadic Alzheimer’s Disease (sAD)
Most studies have focused in obtaining models of organoids based on fAD mutations rather than sAD, in part, because studies conducted on sAD hCOs have observed that they take longer to present associated histopathology [113]. Even with this handicap of time, this type of model can be useful when recapitulating the AD phenotype, such as acceleration of maturation and synapses, increase in the Aβ42/Aβ40 ratio, increase in p-Tau [19, 114, 115], as well as the increase in lipid droplet [115] allowing studying the alterations that trigger AD without being subject to mutations of fAD.
These hCOs sAD models have also laid the foundations for the generation of platforms that allow us to detect new useful drugs against AD. This is because apart from collecting the AD phenotype, they also allow studying drug penetration, as well as studying alterations in numerous pathways: axon development, platelet aggregation, RNA translation, inflammatory ions [116, 117] when comparing the spheroids with post-mortem brains from AD patients. Platforms that are more recent have used organoids with APOE3 and APOE4 identity together with RNA-seq, calcium, and protein quantification analysis against controls to establish a mathematical platform for more realistic drug testing [118].
The investigations carried out on hCOs originating from lines of sAD patients, in contrast to the studies carried out with fAD organoids and, as already mentioned, with their use to generate drug testing platforms, have been considered from the perspective of obtaining possible drugs to alleviate symptomatology. An example would be its use to learn more about tauopathies. Based on the 2D culture background where a relationship between Tau and acetylated histone 6 (HDAC6) was observed [119], the use of the compound CKD-504, a selective and dose-dependent anti-HDAC6, has been studied on these hCOs, obtaining a reduction in p-Tau levels [120].
This use of hCOs to search for alternatives to alleviate the disease has also occurred in organoids obtained from lines with the main risk factor in sAD, the APOE gene. Mainly, lines with APOEε4/ε4 have been used for this purpose, as they are the most aggressive and the only isoform where p-Tau and lipid droplet in hCOS can be seen [19, 114, 115, 118], as well as APOEε3/ε3. Some factors, such as the repressor element silencing transcription factor (REST) 1, have been identified as delaying the onset of symptoms in hCOs APOE4 [114]. A reduction in symptoms has also been observed when APOE4 lines are converted to APOE3 using CRISPR/CAS9 techniques [19, 113], although there are authors who consider that these differences are due more to variations in the culture of hCOS than in the isophorm used for its generation [121].
Despite the lack of depth on how APOE affects to trigger AD [122], its relationship in the loss of typical synapses in AD is known, by acting on the synaptic protein α-synuclein [123] as well as its importance in lipid control, mainly at the neuronal level [115, 123]. However, APOE is not the only gene of interest in sAD. Recent studies are beginning to focus on other genes that might predispose to AD. This is the case of bridging integrator (BIN), which influences the size of the primary endosomes, depending on the isoform present, with isoform 1 (BIN1) being highly associated with early alterations in AD [124]. Likewise, another gene whose alteration is known to affect the correct degradation of Aβ in mitochondria and alters the potential of the mitochondrial membrane is PITRM1 [125]. Studies on hCOs PITRM1 knockout showed AD-like pathology: protein aggregates, tau pathology, and cell death. Mitochondrial alteration was also appreciated, as well as deregulation of astrocytes [126].
Apart from these, various risk factors for sAD, certain studies have considered focusing on the risks of developing sAD due to viral infections. It has been observed that we start from healthy lines infected with HSV-1 [127, 128], or from lines of AD patients infected with the zika virus [129], organoids develop with an increased AD phenotype. Another virus that could have implications for AD is SARS-CoV-2, as it has been associated with destruction of microglia [130], as well as infection of cells with the APOE4 phenotype [72]. Even though it can be considered a way to model AD, using antivirals results in a decrease in AD-like [127, 128], so they end up being a realistic model of the pathology. In addition to viruses, also bacterial infections could influence the onset of AD. It has been observed that in these infections there is an increase in the Aβ level, since it would have a protective role against antimicrobials [131].
3.2.3. Other Approaches to the Study of AD
Another way to model AD without using fAD or sAD patient lines is by inducing the disease from healthy lines. A factor that allows this modulation is aftin-5, which when used on healthy hCOs, it is observed that produces an increase in Aβ42, which is also modulable [132]. Another alternative is based on inducing the pathology by stimulating them with Aβ [133, 134] or with serum from AD patients, which makes it possible to observe, in addition to Aβ and Tau aggregates, neuronal loss and other alterations [135].
Apart from inducing the pathology with various factors, we can submit healthy lines to CRISPR/Cas9 genomic editing or through the use of vectors, so that they express AD. Following these methods, hCOs have been developed in a way that allows us to study the tauopathies [136, 137]. Tau is observed to appear early in human brain development and the concentration of its mRNA is greatly increased in mature neurons [138], for this reason, it is sought how to counteract its effects. For example, p25, a proteolytic fragment of the p35 regulatory subunit, has been shown to induce aberrant cyclin-dependent kinase 5 (Cdk5) activity, which is associated with neurodegenerative disorders such as AD. In organoids, in which Tau effects were induced, p25 blockade was shown to reduce p-Tau levels and increase synaptophysin expression [136]. This type of model allows us to focus on specific causes of AD for a preliminary study where we can simplify the disease.
Although the genes or histopathologies associated with AD are usually studied, some groups have preferred to focus on learning more about genes or factors that are protective against the pathology. One of these genes is BACE2, located on chromosome 21. Using organoids from patients with DS (with an extra copy of APP) and AD (APOE3 phenotype), the effects of BACE2 have been studied. This gene was found to produce high levels of amyloid-free peptides and Aβ degradation products [139]. However, in addition, if we start from a line with mutations in BACE2, the organoids that are generated will present a pathology similar to AD [140]. Similarly, ADAM10, the gene encoding α-secretase, is interesting, since this secretase can cleave APP instead of β-secretase without giving rise to Aβ peptides. In organoids that have been induced for AD by Aβ peptides, it is observed that activation of ADAM10 reduces the pathology. The levels of these peptides can be reduced by adding oestrogens to the medium, since these are involved in ADAM10 activity [133]. Continuing with this type of stimulation to obtain AD in organoids, a decrease in mitophagy has also been observed, as well as one of its associated genes, PINK1. It was observed that this reduction could be alleviated with the addition of a flavonoid called galangin [134].
4. Future Perspectives
Recent advances in the field of cell cultures have opened up a wide range of opportunities in the study of human diseases, making it possible to overcome the disadvantages of previous models. Despite the advantages of animal models, such as their easy genetic manipulation, physiological differences with the human brain have led to the emergence of in vitro 2D models [141]. These 2D cultures allow obtaining highly pure, homogeneous and reproducible cell cultures, but they cannot recreate some neurodevelopmental events, such as cell-scaffold interaction or the influence of some cells on other cell types [34]. For this reason, hCOs have caused a great revolution [142]. Even so, animal models and 2D cultures remain important for preliminary steps in research [104, 127], as they allow the results to be verified at different levels. Also, when it comes to neurodegenerative diseases and age-related disorders, it should be noted that, hESCs and other stem cells have a fetal or embryonic cell age, which is a limitation for these studies [105, 127].
As explained above, in vitro 3D models have compensated for the shortcomings of previous models, providing a more reliable view of cell interaction, drug response, and spatiotemporal development. Other advantages are that it can allow us to obtain organoids from patients for more personalized treatment [82] whole organoid image analyses can be performed for a global understanding [143]. It also allows us to reproduce and study the extracellular environment [100], how to use them for the creation of virtual platforms, allowing us to test new strategies without long protocols or ethical conflicts [118, 144, 145], or delving into stem cell therapy [146]. However, obtaining these organoids requires long-term experiments due to slow growth and the times required for cell maturation, aging and senescence [147].
One of the disadvantages of hCOs is ageing, which is due to hypoxic areas in deep regions due to poor perfusion of nutrients and other substances, as they lack vascularisation [148]. This limits in-depth study of neurodegenerative phenotypes and chemical detection. Several strategies have been proposed to address this. A first option would be to use support materials, such as matrigel, porous structures, hydrogels or polymeric materials [149-151]. Even so, these structures are difficult to use, so improvements are still being sought [152], or not relying on supports for their cultivation [153]. Another option that has been considered is to vascularize hCOs. To this end, it has been proposed to transplant organoids into mice [154, 155], but also to directly generate this vasculature [156-158]. Another option for vascularisation would be through co-culture with cells [159-161]. Studying vascularisation would allow us to learn more about the blood-brain barrier (BBB), whose permeability increases in AD, allowing us to improve drug screening [162]. The last strategy that has been proposed is to reduce senescence [163, 164].
The heterogeneity of organoids is another of its limitations. The use of embryoid bodies is a bottleneck for organoid formation, decreasing efficiency and homogeneity. Other factors that increase their heterogeneity are the culture conditions themselves [121], the AD mutation to be studied [129], the material of the plates where they are cultivated or in which at the time of the protocol, matrigel is used [165].
Another drawback is that, so far, most protocols have failed to recapitulate all of the cell types that make up the brain. Even so, one group had recently created a 2D triculture of neurons, microglia, and astrocytes [166, 167]. This opens up an opportunity to create three-culture organoids for more complex models. For example, it is already possible to obtain spheroids that also include oligodendrocytes [168]. The same as obtaining mixed organoids of neurons and astrocytes by fusing them after culturing each cell type independently [115].
5. Conclusion
As seen throughout this review, 3D culture models, with an emphasis on hCOs, appear to be one of the best models for advancing AD research today. Compared to previous models (in vivo animals and 2D in vitro cultures), hCOs have been found not only to be more efficient, but also to recapitulate some of the most striking features of AD pathology. This has allowed enormous progress in the field in recent years.
Human hCOs, by recapitulating a large part of the complexity of the human brain and due to the innovation of their technology, are quickly positioning themselves in a very interesting model for the study of the molecular and cellular pathology characteristic of AD at the brain level. These hCOs can be generated from control iPS cells or from AD patients, both fAD and sAD, allowing mutations or genes of interest to be introduced or deleted by gene editing. Thus, new therapeutic targets or possible treatments could be determined more quickly.
Another advantage of hCOs is that it is a very suitable technology to further investigate the interactions of the different phenotypes of brain cells (neuron-astrocytes-oligodendrocytes-microglia), under not only physiological conditions, but also pathological ones. Although, there are still challenges to improve and deepen the use of these models, even with the current limitations, great results have already been achieved, allowing for a better understanding of AD, as well as advances in potential therapies.
Funding
This research was supported grant number PID2021-126715OB-I00 funded by MCIN/AEI/10.13039/ 501100011033 and by “ERDF A way of making Europe” and by the Grant of Instituto de Salud Carlos III (ISCIII), AESI, PI22CIII/00055.
Acknowledgments
The authors thank to Laura Maeso for contributing in the processing of the review of the manuscript.
Conflicts of Interest
None.
REFERENCES
[1]
Alzheimer's Association “2013 Alzheimer's disease
facts and figures..” Alzheimers Dement, vol. 9, no. 2, pp. 208-245,
2013. View at: Publisher
Site | PubMed
[2]
Henry Brodaty, Monique M B Breteler, et al. “The World
of Dementia beyond 2020.” J Am Geriatr Soc, vol. 59, no. 5, pp. 923-927,
2011. View at: Publisher
Site | PubMed
[3]
Sergio T Ferreira, Mychael V Lourenco, Mauricio M
Oliveira, et al. “Soluble Amyloid-β Oligomers as Synaptotoxins Leading to Cognitive Impairment in Alzheimer’s Disease.” Front
Cell Neurosci, vol. 9, pp. 191, 2015. View at: Publisher
Site | PubMed
[4]
Divya Goyal, Syed Afroz Ali, Rakesh Kumar Singh
“Emerging Role of Gut Microbiota in Modulation of Neuroinflammation and
Neurodegeneration with Emphasis on Alzheimer’s Disease.” Prog
Neuropsychopharmacol Biol Psychiatry, vol. 106, pp. 110112, 2021. View at: Publisher
Site | PubMed
[5]
Richard Mayeux, Yaakov Stern “Epidemiology of
Alzheimer Disease.” Cold Spring Harb Perspect Med, vol. 2, no. 8, pp.
a006239, 2012. View at: Publisher
Site | PubMed
[6]
Leslie Sibener, Ibrahim Zaganjor, Heather M Snyder, et
al. “Alzheimer’s Disease Prevalence, Costs, and Prevention for Military
Personnel and Veterans.” Alzheimers Dement, vol. 10, no. 3 Suppl, pp.
S105-S110, 2014. View at: Publisher Site | PubMed
[7]
Jianping Jia, Cuibai Wei, Shuoqi Chen, et al. “The
Cost of Alzheimer’s Disease in China and Re-Estimation of Costs Worldwide.”
Alzheimers Dement, vol. 14, no. 4, pp. 483-491, 2018. View at: Publisher
Site | PubMed
[8]
Takashi Amemori, Pavla Jendelova, Jiri Ruzicka, et al.
“Alzheimer’s Disease: Mechanism and Approach to Cell Therapy.” Int J Mol Sci,
vol. 16, no. 11, pp. 26417-26451, 2015. View at: Publisher Site | PubMed
[9]
Alistair Burns, E Jane Byrne, Konrad Maurer “Alzheimer
’s disease.” Lancet, vol. 360, no. 9327, pp. 163-165, 2002. View at: Publisher
Site | PubMed
[10]
Jay Penney, William T Ralvenius, Li-Huei Tsai
“Modeling Alzheimer’s Disease with IPSC-Derived Brain Cells.” Mol Psychiatry,
vol. 25, no. 1, pp. 148-167, 2020. View at: Publisher Site | PubMed
[11]
Jose A Soria Lopez, Hector M González,
Gabriel C Léger “Alzheimer’s Disease.” Handb Clin Neurol, vol. 167,
pp. 231-255, 2019. View at: Publisher Site | PubMed
[12]
Joel W Blanchard, Matheus B Victor, Li-Huei Tsai
“Dissecting the Complexities of Alzheimer Disease with in vitro Models
of the Human Brain.” Nat Rev Neurol, vol. 18, no. 1, pp. 25-39, 2022.
View at: Publisher
Site | PubMed
[13]
Alberto Serrano-Pozo, Matthew P Frosch, Eliezer
Masliah, et al. “Neuropathological Alterations in Alzheimer Disease.” Cold
Spring Harb Perspect Med, vol. 1, no. 1, pp. a006189, 2011. View at: Publisher
Site | PubMed
[14]
Marc Aurel Busche, Bradley T Hyman “Synergy between
Amyloid-β and Tau in
Alzheimer’s Disease.” Nat Neurosci, vol. 23, no. 10, pp. 1183-1193,
2020. View at: Publisher
Site | PubMed
[15]
Huiqin Zhang, Wei Wei 1, Ming Zhao, et al.
“Interaction between Aβ and Tau in
the Pathogenesis of Alzheimer’s Disease.” Int J Biol Sci, vol. 17, no. 9, pp. 2181-2192, 2021. View at: Publisher Site | PubMed
[16]
Jana Povova, Petr Ambroz, Michal Bar, et al.
“Epidemiological of and Risk Factors for Alzheimer’s Disease: A Review.” Biomed
Pap Med Fac Univ Palacky Olomouc Czech Repub, vol. 156, no. 2, pp. 108-114,
2012. View at: Publisher
Site | PubMed
[17]
Caroline Van Cauwenberghe, Christine Van Broeckhoven,
Kristel Sleegers “The Genetic Landscape of Alzheimer Disease: Clinical
Implications and Perspectives.” Genet Med, vol. 18, no. 5, pp. 421-430,
2016. View at: Publisher
Site | PubMed
[18]
Alexei Verkhratsky, Markel Olabarria, Harun N
Noristani, et al. “Astrocytes in Alzheimer’s Disease.” Neurotherapeutics, vol.
7, no. 4, pp. 399-412, 2010. View at: Publisher Site | PubMed
[19]
Jing Zhao, Yuan Fu 1, Yu Yamazaki, et al. “APOE4
Exacerbates Synapse Loss and Neurodegeneration in Alzheimer’s Disease Patient
IPSC-Derived Cerebral Organoids.” Nat Commun, vol. 11, no. 1, 2020. View
at: Publisher
Site | PubMed
[20]
Lynn M Bekris, Chang-En Yu, Thomas D Bird, et al.
“Genetics of Alzheimer Disease.” J Geriatr Psychiatry Neurol, vol. 23,
no. 4, pp. 213-227, 2010. View at: Publisher Site | PubMed
[21]
Jeffrey L Cummings, Gary Tong, Clive Ballard
“Treatment Combinations for Alzheimer’s Disease: Current and Future Pharmacotherapy
Options.” J Alzheimers Dis, vol. 67, no. 3, pp. 779-794, 2019.View at: Publisher Site | PubMed
[22]
Anil Kumar, Arti Singh, Ekavali “A Review on
Alzheimer’s Disease Pathophysiology and Its Management: An Update.” Pharmacol
Rep, vol. 67, no.2, pp. 195-203, 2015. View at: Publisher
Site | PubMed
[23]
Juan Yang, Song Li, Xi-Biao He, et al. “Induced
Pluripotent Stem Cells in Alzheimer’s Disease: Applications for Disease
Modeling and Cell-Replacement Therapy.” Mol Neurodegener, vol. 11, no.
1, pp. 39, 2016. View at: Publisher Site | PubMed
[24]
Joel Petch, Daniel Bressington “Aducanumab for
Alzheimer’s Disease: The Never-Ending Story That Nurses Should Know.” Nurs
Open, vol. 8, no. 4, pp. 1524-1526, 2021. View at: Publisher Site | PubMed
[25]
Stephen Salloway, Spyros Chalkias, Frederik Barkhof,
et al. “Amyloid-Related Imaging Abnormalities in 2 Phase 3 Studies Evaluating
Aducanumab in Patients with Early Alzheimer Disease.” JAMA Neurol, vol.
79, no. 1, pp. 13-21, 2022. View at: Publisher Site | PubMed
[26]
Eleanor Drummond, Thomas Wisniewski “Alzheimer’s
Disease: Experimental Models and Reality.” Acta Neuropathol, vol. 133,
no. 2, pp. 155-175, 2017. View at: Publisher Site | PubMed
[27]
Tarja Malm, Johanna Magga, Jari Koistinaho “Animal
Models of Alzheimer’s Disease: Utilization of Transgenic Alzheimer’s Disease
Models in Studies of Amyloid Beta Clearance.” Curr Transl Geriatr Exp
Gerontol Rep, vol. 1, no. 1, pp. 11-20, 2012. View at: Publisher
Site | PubMed
[28]
Tian Qin, Samantha Prins, Geert Jan Groeneveld, et al.
“Utility of Animal Models to Understand Human Alzheimer’s Disease, Using the
Mastermind Research Approach to Avoid Unnecessary Further Sacrifices of
Animals.” Int J Mol Sci, vol. 21, no. 9, pp. 3158, 2020. View at: Publisher
Site | PubMed
[29]
Tara L Spires, Bradley T Hyman “Transgenic Models of
Alzheimer’s Disease: Learning from Animals.” NeuroRx, vol. 2, no. 3, pp.
423-437, 2005. View at: Publisher
Site | PubMed
[30]
Mason A Israel 1, Shauna H Yuan, Cedric Bardy, et al.
“Probing Sporadic and Familial Alzheimer’s Disease Using Induced Pluripotent
Stem Cells.” Nature, vol. 482, no. 7384, pp. 216-220, 2012. View at: Publisher
Site | PubMed
[31]
Simona Baldassari, Ilaria Musante, Michele Iacomino,
et al. “Brain Organoids as Model Systems for Genetic Neurodevelopmental
Disorders.” Front Cell Dev Biol, vol. 8, pp. 590119, 2020. View at: Publisher
Site | PubMed
[32]
Juliana Minardi Nascimento, Verônica M Saia-Cereda,
Rafaela C Sartore, et al. “Human Cerebral Organoids and Fetal Brain Tissue
Share Proteomic Similarities.” Front Cell Dev Biol, vol. 7, pp. 303,
2019. View at: Publisher
Site | PubMed
[33]
Zizhen Si, Xidi Wang “Stem Cell Therapies in
Alzheimer’s Disease: Applications for Disease Modeling.” J Pharmacol Exp
Ther, vol. 377, no. 2, pp. 207-217, 2021. View at: Publisher
Site | PubMed
[34]
Hsih-Yin Tan, Hansang Cho, Luke P Lee “Human
Mini-Brain Models.” Nat Biomed Eng, vol. 5, no, 1, pp. 11-25, 2021. View
at: Publisher
Site | PubMed
[35]
Francesca Pistollato, Elan L Ohayon, Ann Lam, et al.
“Alzheimer Disease Research in the 21 St Century: Past and Current Failures,
New Perspectives and Funding Priorities.” Oncotarget, vol. 7, no. 26.
pp. 38999-39016, 2016. View at: Publisher Site | PubMed
[36]
Se Hoon Choi, Young Hye Kim, Luisa Quinti, et al. “3D
Culture Models of Alzheimer’s Disease: A Road Map to a “Cure-in-a-Dish”.” Mol
Neurodegener, vol. 11, no. 1, pp. 75, 2016. View at: Publisher
Site | PubMed
[37]
Godwin Tong, Pablo Izquierdo, Rana Arham Raashid
“Human Induced Pluripotent Stem Cells and the Modelling of Alzheimer’s Disease:
The Human Brain Outside the Dish.” Open Neurol J, vol. 11, pp. 27-38,
2017. View at: Publisher
Site | PubMed
[38]
Silvia Benito-Kwiecinski, Stefano L Giandomenico,
Magdalena Sutcliffe, et al. “An Early Cell Shape Transition Drives Evolutionary
Expansion of the Human Forebrain.” Cell, vol. 184, no. 8, pp. 2084-2102,
2021. View at: Publisher
Site | PubMed
[39]
Sandra J Engle, Laura Blaha, Robin J Kleiman “Best
Practices for Translational Disease Modeling Using Human IPSC-Derived Neurons.”
Neuron, vol. 100, no. 4, pp. 783-797, 2018. View at: Publisher
Site | PubMed
[40]
Kazutoshi Takahashi 1, Koji Tanabe, Mari Ohnuki, et
al. “Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by
Defined Factors.” Cell, vol. 131, no. 5, pp. 861-872, 2007. View at: Publisher
Site | PubMed
[41]
George Q Daley “Stem Cells and the Evolving Notion of
Cellular Identity.” Philos Trans R Soc Lond B Biol Sci, vol. 370,, no.
1680, 2015. View at: Publisher
Site | PubMed
[42]
Andreea Rosca, Raquel Coronel, Miryam Moreno, et al.
“Impact of Environmental Neurotoxic: Current Methods and Usefulness of Human
Stem Cells.” Heliyon, vol. 6, no. 12, pp. e05773, 2020. View at: Publisher
Site | PubMed
[43]
Junying Yu, Maxim A Vodyanik, Kim Smuga-Otto, et al.
“Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells.” Science,
vol. 318, no. 5858, pp. 1917-1920, 2007. View at: Publisher
Site | PubMed
[44]
Nasir Malik, Mahendra S Rao “A Review of the Methods
for Human IPSC Derivation.” Methods Mol Biol, vol. 997, pp. 23-33, 2013.
View at: Publisher
Site | PubMed
[45]
Kazutoshi Takahashi, Shinya Yamanaka “A Decade of
Transcription Factor-Mediated Reprogramming to Pluripotency.” Nat Rev Mol
Cell Biol, vol. 17, no. 3, pp. 183-193, 2016. View at: Publisher Site | PubMed
[46]
Vasiliki Mahairaki, Jiwon Ryu, Ann Peters, et al.
“Induced Pluripotent Stem Cells from Familial Alzheimer’s Disease Patients
Differentiate into Mature Neurons with Amyloidogenic Properties.” Stem Cells
Dev, vol. 23, no. 24, pp. 2996-3010, 2014. View at: Publisher
Site | PubMed
[47]
Takuya Yagi 1, Daisuke Ito, Yohei Okada, et al.
“Modeling Familial Alzheimer’s Disease with Induced Pluripotent Stem Cells.”
Hum Mol Genet, vol. 20, no. 23, pp. 4530-4539, 2011. View at: Publisher Site | PubMed
[48]
Naoki Yahata, Masashi Asai, Shiho Kitaoka, et al.
“Anti-Aβ Drug
Screening Platform Using Human IPS Cell-Derived Neurons for the Treatment of
Alzheimer’s Disease.” PLoS One,
vol. 6, no. 9, 2011. View at: Publisher Site | PubMed
[49]
Amir M Hossini, Matthias Megges, Alessandro Prigione,
et al. “Induced Pluripotent Stem Cell-Derived Neuronal Cells from a Sporadic
Alzheimer’s Disease Donor as a Model for Investigating AD-Associated Gene
Regulatory Networks.” BMC Genomics, vol. 16, no. 1, 2015. View at: Publisher
Site | PubMed
[50]
Grace Woodruff 1, Jessica E Young, Fernando J
Martinez, et al. “The Presenilin-1 ΔE9 Mutation Results in Reduced γ-Secretase Activity, but Not Total Loss of PS1 Function, in Isogenic
Human Stem Cells.” Cell Rep,
vol. 5, no. 4, pp. 974-985, 2013. View at: Publisher Site | PubMed
[51]
Steven Moore, Lewis D B Evans, Therese Andersson, et
al. “APP Metabolism Regulates Tau Proteostasis in Human Cerebral Cortex
Neurons.” Cell Rep, vol. 11, no. 5, pp. 689-696, 2015. View at: Publisher
Site | PubMed
[52]
Sean Schrank, John McDaid, Clark A Briggs, et al.
“Human-Induced Neurons from Presenilin 1 Mutant Patients Model Aspects of
Alzheimer’s Disease Pathology.” Int J Mol Sci, vol. 21, no. 3, 2020.
View at: Publisher
Site | PubMed
[53]
Minna Oksanen, Andrew J Petersen, Nikolay Naumenko, et
al. “PSEN1 Mutant IPSC-Derived Model Reveals Severe Astrocyte Pathology in
Alzheimer’s Disease.” Stem Cell Reports, vol. 9, no. 6, pp. 1885-1897,
2017. View at: Publisher
Site | PubMed
[54]
Juan Beauquis, Angeles Vinuesa, Carlos Pomilio, et al.
“Hippocampal and Cognitive Alterations Precede Amyloid Deposition in a Mouse
Model for Alzheimer’s Disease.” Medicina (B Aires), vol. 74, no. 4, pp.
282-286, 2013. View at: PubMed
[55]
V C Jones, R Atkinson-Dell, A Verkhratsky, et al.
“Aberrant IPSC-Derived Human Astrocytes in Alzheimer’s Disease.” Cell Death
Dis, vol. 8, no. 3, 2017. View at: Publisher Site | PubMed
[56]
Takayuki Kondo, Masashi Asai, Kayoko Tsukita, et al. “Modeling
Alzheimer’s Disease with Reveals Stress Phenotypes Associated with
Intracellular Aβ and
Differential Drug Responsiveness.” Cell Stem
Cell, vol. 12, no. 4, pp. 487-496, 2013. View at: Publisher
Site | PubMed
[57]
Christina R Muratore, Heather C Rice, Priya Srikanth,
et al. “The Familial Alzheimer’s Disease APPV717I Mutation Alters APP
Processing and Tau Expression in IPSC-Derived Neurons.” Hum Mol Genet,
vol. 23, no. 13, pp. 3523-3536, 2014. View at: Publisher Site | PubMed
[58]
Lauren K Fong, Max M Yang, Rodrigo Dos Santos Chaves,
et al. “Full-Length Amyloid Precursor Protein Regulates Lipoprotein Metabolism
and Amyloid- Clearance in Human Astrocytes.” J Biol Chem, vol. 293, ,
no. 29, pp. 11341-11357, 2018. View at: Publisher Site | PubMed
[59]
Tânia Fernandes, Rosa Resende, Diana F Silva, et al.
“Structural and Functional Alterations in Mitochondria-Associated Membranes
(Mams) and in Mitochondria Activate Stress Response Mechanisms in an In
Vitro Model of Alzheimer’s Disease.” Biomedicines, vol. 9, no. 8,
pp. 881, 2021. View at: Publisher
Site | PubMed
[60]
Rik van der Kant, Vanessa F Langness, Cheryl M
Herrera, et al. “Cholesterol Metabolism Is a Druggable Axis That Independently
Regulates Tau and Amyloid-β in
IPSC-Derived Alzheimer’s Disease
Neurons.” Cell Stem Cell, vol. 24, no. 3, pp.
363-375, 2019. View at: Publisher
Site | PubMed
[61] M Catarina
Silva, Ghata A Nandi, Sharon Tentarelli, et al. “Prolonged Tau Clearance and
Stress Vulnerability Rescue by Pharmacological Activation of Autophagy in Tauopathy
Neurons.” Nat Commun, vol. 11,
no. 1, pp. 3258, 2020. View at: Publisher Site | PubMed
[62]
Raquel Coronel, María Lachgar, Adela
Bernabeu-Zornoza, et al. “Neuronal and Glial Differentiation of
Human Neural Stem Cells Is Regulated by Amyloid Precursor Protein (APP)
Levels.” Mol Neurobiol, vol. 56, no. 2, pp. 1248-1261, 2019. View at: Publisher
Site | PubMed
[63]
Yichen Shi, Peter Kirwan, James Smith, et al. “A Human
Stem Cell Model of Early Alzheimer’s Disease Pathology in down Syndrome.” Sci
Transl Med, vol. 4, no. 124, pp. 124ra29, 2012. View at: Publisher
Site | PubMed
[64]
Mark W Bondi, Emily C Edmonds, David P Salmon
“Alzheimer’s Disease: Past, Present, and Future.” J Int Neuropsychol Soc,
vol. 23, no. 9-10, pp. 818-831, 2017. View at: Publisher Site | PubMed
[65]
Chia-Yu Chang, Sheng-Mei Chen, Huai-En Lu, et al.
“N-Butylidenephthalide Attenuates Alzheimer’s Disease-like Cytopathy in down
Syndrome Induced Pluripotent Stem Cell-Derived Neurons.” Sci Rep, vol.
5, pp. 8744, 2015. View at: Publisher
Site | PubMed
[66]
Dmitry A Ovchinnikov, Othmar Korn, Isaac Virshup, et
al. “The Impact of APP on Alzheimer-like Pathogenesis and Gene Expression in
Down Syndrome IPSC-Derived Neurons.” Stem Cell Reports, vol. 11, no. 1,
pp. 32-42, 2018. View at: Publisher Site | PubMed
[67]
Christopher J Bissonnette, Ljuba Lyass, Bula J
Bhattacharyya, et al. “The Controlled Generation of Functional Basal Forebrain Cholinergic
Neurons from Human Embryonic Stem Cells.” Stem Cells, vol. 29, no. 5,
pp. 802-811, 2011. View at: Publisher
Site | PubMed
[68]
Lishu Duan, Bula J Bhattacharyya, Abdelhak Belmadani,
et al. “Stem Cell Derived Basal Forebrain Cholinergic Neurons from Alzheimer’s
Disease Patients Are More Susceptible to Cell Death.” Mol Neurodegener,
vol. 9, pp. 3, 2014. View at: Publisher
Site | PubMed
[69]
Jing Zhao, Mary D Davis, Yuka A Martens, et al. “APOE Ε4/Ε4 Diminishes Neurotrophic Function of Human IPSC-Derived Astrocytes.”
Hum Mol Genet, vol. 26, no. 14, pp. 2690-2700, 2017. View at: Publisher Site | PubMed
[70]
Alicia A Nugent, Karin Lin, Bettina van Lengerich, et
al. “TREM2 Regulates Microglial Cholesterol Metabolism upon Chronic Phagocytic
Challenge.” Neuron, vol. 105, no. 5, pp. 837-854, 2020. View at: Publisher
Site | PubMed
[71]
Sreedevi Raman, Gayathri Srinivasan, Nicholas
Brookhouser, et al. “A Defined and Scalable Peptide-Based Platform for the
Generation of Human Pluripotent Stem Cell-Derived Astrocytes.” ACS Biomater
Sci Eng, vol. 6, no. 6, pp. 3477-3490, 2020. View at: Publisher
Site | PubMed
[72]
Cheng Wang 1, Mingzi Zhang 1, Gustavo Garcia Jr, et
al. “ApoE-Isoform-Dependent SARS-CoV-2 Neurotropism and Cellular Response.” Cell
Stem Cell, vol. 28, no. 2, pp. 331-342, 2021. View at: Publisher
Site | PubMed
[73]
Andrew A Sproul, Samson Jacob, Deborah Pre, et al.
“Characterization and Molecular Profiling of PSEN1 Familial Alzheimer’s Disease
IPSC-Derived Neural Progenitors.” PLoS One, vol. 9, no. 1, 2014. View
at: Publisher
Site | PubMed
[74]
Julian H Birnbaum, Debora Wanner, Anton F Gietl, et
al. “Oxidative Stress and Altered Mitochondrial Protein Expression in the
Absence of Amyloid-β and Tau
Pathology in IPSC-Derived Neurons from Sporadic Alzheimer’s Disease Patients.” Stem Cell Res, vol. 27, pp. 121-130, 2018.
View at: Publisher
Site | PubMed
[75]
Noemi Esteras, Franziska Kundel, Giuseppe F Amodeo, et
al. “Insoluble Tau Aggregates Induce Neuronal Death through Modification of
Membrane Ion Conductance, Activation of Voltage-Gated Calcium Channels and
NADPH Oxidase.” FEBS J, vol. 288, no. 1, pp. 127-141, 2021. View at: Publisher Site | PubMed
[76]
Hazel Hall-Roberts, Devika Agarwal, Juliane Obst, et
al. “TREM2 Alzheimer’s Variant R47H Causes Similar Transcriptional
Dysregulation to Knockout, yet Only Subtle Functional Phenotypes in Human
IPSC-Derived Macrophages.” Alzheimers Res Ther, vol. 12, no. 1, 2020.
View at: Publisher
Site | PubMed
[77]
Zhiqiang Liu, Carlo Condello, Aaron Schain, et al.
“CX3CR1 in Microglia Regulates Brain Amyloid Deposition through Selective
Protofibrillar Amyloid-β
Phagocytosis.” J Neurosci, vol. 30, no. 50, pp.
17091-17101, 2010. View at: Publisher Site | PubMed
[78]
Edsel M Abud, Ricardo N Ramirez, Eric S Martinez, et
al. “IPSC-Derived Human Microglia-like Cells to Study Neurological Diseases.” Neuron,
vol. 94, no. 2, pp. 278-293, 2017. View at: Publisher Site | PubMed
[79]
Evandro F Fang, Yujun Hou, Konstantinos Palikaras, et
al. “Mitophagy Inhibits Amyloid-β and Tau Pathology and Reverses Cognitive Deficits in Models of
Alzheimer’s Disease.” Nat
Neurosci, vol. 22, no. 3, pp. 401-412, 2019. View at: Publisher
Site | PubMed
[80]
Russell H Swerdlow “Mitochondria and Mitochondrial
Cascades in Alzheimer’s Disease.” J Alzheimers Dis, vol. 62, no. 3, pp.
1403-1416, 2018. View at: Publisher
Site | PubMed
[81]
William A Banks, Joshua B Owen, Michelle A Erickson
“Insulin in the Brain: There and Back Again.” Pharmacol Ther, vol. 136, no. 1,
pp. 82-93, 2012. View at: Publisher Site | PubMed
[82]
Yujung Chang, Junyeop Kim 1, Hanseul Park, et al.
“Modelling Neurodegenerative Diseases with 3D Brain Organoids.” Biol Rev
Camb Philos Soc, vol. 95, no. 5, pp. 1497-1509, 2020. View at: Publisher Site | PubMed
[83]
Gonçalo Garcia, Sara Pinto, Mar Cunha, et al.
“Neuronal Dynamics and Mirna Signaling Differ between Sh-Sy5y Appswe and Psen1
Mutant Ipsc-Derived Ad Models upon Modulation with Mir-124 Mimic and
Inhibitor.” Cells, vol. 10, no. 9, 2021. View at: Publisher
Site | PubMed
[84]
Madeline A Lancaster, Magdalena Renner, Carol-Anne
Martin, et al. “Cerebral Organoids Model Human Brain Development and
Microcephaly.” Nature, vol. 501, no. 7467, pp. 373-379, 2013. View at: Publisher
Site | PubMed
[85]
Madeline A Lancaster, Juergen A Knoblich
“Organogenesisin a Dish: Modeling Development and Disease Using Organoid
Technologies.” Science, vol. 345, no. 6194, pp. 1247125, 2014. View at: Publisher
Site | PubMed
[86]
Giorgia Quadrato, Juliana Brown, Paola Arlotta “The
Promises and Challenges of Human Brain Organoids as Models of Neuropsychiatric
Disease.” Nat Med, vol. 22, no. 11, pp. 1220-1228, 2016. View at: Publisher Site | PubMed
[87]
Laleh Shariati, Yasaman Esmaeili, Shaghayegh Haghjooy
Javanmard, et al. “Organoid Technology: Current Standing and Future
Perspectives.” Stem Cells, vol. 39, no. 12, pp. 1625-1649, 2021. View
at: Publisher Site | PubMed
[88]
Yoshiki Sasai “Next-Generation Regenerative Medicine:
Organogenesis from Stem Cells in 3D Culture.” Cell Stem Cell, vol. 12,
no. 5, pp. 520-530, 2013. View at: Publisher Site | PubMed
[89]
Steven A Sloan, Jimena Andersen, Anca M Pașca, et al.
“Generation and Assembly of Human Brain Region-Specific Three-Dimensional
Cultures.” Nat Protoc, vol. 13, no. 9, pp. 2062-2085, 2018. View at: Publisher
Site | PubMed
[90]
Anca M Paşca, Steven A Sloan, Laura E Clarke, et al. “Functional Cortical Neurons
and Astrocytes from Human Pluripotent Stem Cells in 3D Culture.” Nat Methods,
vol. 12, no. 7, pp. 671-678, 2015. View at: Publisher Site | PubMed
[91]
Yangfei Xiang, Yoshiaki Tanaka, Benjamin Patterson, et
al. “Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain
Development and Interneuron Migration.” Cell Stem Cell, vol. 21, no. 3,
pp. 383-398, 2017. View at: Publisher Site | PubMed
[92]
Ian T Fiddes, Gerrald A Lodewijk, Meghan Mooring, et
al. “Human-Specific NOTCH2NL Genes Affect Notch Signaling and Cortical
Neurogenesis.” Cell, vol. 173, no. 6, pp. 1356-1369, 2018. View at: Publisher
Site | PubMed
[93]
Mayur Madhavan, Zachary S Nevin, H Elizabeth Shick, et
al. “Induction of Myelinating Oligodendrocytes in Human Cortical Spheroids.” Nat
Methods, vol. 15, no. 9, pp. 700-706, 2018. View at: Publisher
Site | PubMed
[94]
Cleber A Trujillo, Richard Gao, Priscilla D Negraes,
et al. “Complex Oscillatory Waves Emerging from Cortical Organoids Model Early
Human Brain Network Development.” Cell Stem Cell, vol. 25, no. 4, pp.
558-569, 2019. View at: Publisher
Site | PubMed
[95]
Silvia Velasco, Amanda J Kedaigle, Sean K Simmons, et
al. “Individual Brain Organoids Reproducibly Form Cell Diversity of the Human
Cerebral Cortex.” Nature, vol. 570, no. 7762, pp. 523-527, 2019. View
at: Publisher
Site | PubMed
[96]
J Gray Camp, Farhath Badsha, Marta Florio, et al.
“Human Cerebral Organoids Recapitulate Gene Expression Programs of Fetal
Neocortex Development.” Proc Natl Acad Sci U S A, vol. 112, no. 51, pp.
15672-15677, 2015. View at: Publisher
Site | PubMed
[97]
Waseem K Raja, Alison E Mungenast, Yuan-Ta Lin, et al.
“Self-Organizing 3D Human Neural Tissue Derived from Induced Pluripotent Stem
Cells Recapitulate Alzheimer’s Disease Phenotypes.” PLoS One, vol. 11,
no. 9, pp. e0161969, 2016. View at: Publisher Site | PubMed
[98]
Hélène-Marie Lanoiselée, Gaël Nicolas, David Wallon,
et al. “APP, PSEN1, and PSEN2 Mutations in Early-Onset Alzheimer
Disease: A Genetic Screening Study of Familial and Sporadic Cases.” PLoS Med,
vol. 14, no. 3, pp. e1002270, 2017. View at: Publisher Site | PubMed
[99]
HyunJung Park, Jaehyeon Kim, Chongsuk Ryou “A
Three-Dimensional Spheroid Co-Culture System of Neurons and Astrocytes Derived
from Alzheimer’s Disease Patients for Drug Efficacy Testing.” Cell Prolif,
vol. 56, no. 6, pp. e13300, 2023. View at: Publisher Site | PubMed
[100]
Yuanwei Yan, Liqing Song, Julie Bejoy, et al.
“Modeling Neurodegenerative Microenvironment Using Cortical Organoids Derived
from Human Stem Cells.” Tissue Eng Part A, vol. 24, no. 13-14, pp.
1125-1137, 2018. View at: Publisher Site | PubMed
[101]
Nam Gyo Kim, Dong Ju Jung, Yeon-Kwon Jung, et al. “The
Effect of a Novel Mica Nanoparticle, STB-MP, on an Alzheimer’s Disease
Patient-Induced PSC-Derived Cortical Brain Organoid Model.” Nanomaterials
(Basel), vol. 13, no. 5, pp. 893, 2023. View at: Publisher
Site | PubMed
[102]
Hiroko Shimada, Yuta Sato, Takashi
Sasaki, et al. “A Next-Generation IPSC-Derived Forebrain Organoid
Model of Tauopathy with Tau Fibrils by AAV-Mediated Gene Transfer.” Cell Rep
Methods, vol. 2, no. 9, pp. 100289, 2022. View at: Publisher
Site | PubMed
[103]
Juan Yin, Antonius M VanDongen “Enhanced Neuronal
Activity and Asynchronous Calcium Transients Revealed in a 3D Organoid Model of
Alzheimer’s Disease.” ACS Biomater Sci Eng, vol. 7, no. 1, pp. 254-264,
2021. View at: Publisher
Site | PubMed
[104]
Charles Arber, Jamie Toombs, Christopher Lovejoy, et
al. “Familial Alzheimer’s Disease Patient-Derived Neurons Reveal Distinct
Mutation-Specific Effects on Amyloid Beta.” Mol Psychiatry, vol. 25, no.
11, pp. 2919-2931, 2020. View at: Publisher Site | PubMed
[105]
Swagata Ghatak, Nima Dolatabadi, Dorit Trudler, et al.
“Mechanisms of Hyperexcitability in Alzheimer’s Disease hiPSC-Derived Neurons
and Cerebral Organoids vs. Isogenic Control.” Elife, vol. 8, pp. e50333,
2019. View at: Publisher
Site | PubMed
[106]
Charles Arber, Christopher Lovejoy, Lachlan Harris, et
al. “Familial Alzheimer’s Disease Mutations in PSEN1 Lead to Premature
Human Stem Cell Neurogenesis.” Cell Rep, vol. 34, no. 2, 2021. View at: Publisher
Site | PubMed
[107]
Erin M Hurley, Pawel Mozolewski, Radek Dobrowolski, et
al. “Familial Alzheimer’s Disease-Associated PSEN1 Mutations Affect
Neurodevelopment through Increased Notch Signaling.” Stem Cell Reports,
vol. 18, no. 7, pp. 1516-1533, 2023. View at: Publisher Site | PubMed
[108]
Swagata Ghatak, Nima Dolatabadi, Richard Gao, et al.
“NitroSynapsin Ameliorates Hypersynchronous Neural Network Activity in
Alzheimer hiPSC Models.” Mol Psychiatry, vol. 26, no. 10, pp. 5751-5765,
2021. View at: Publisher
Site | PubMed
[109]
Yuriy Pomeshchik, Oxana Klementieva, Jeovanis Gil, et
al. “Human IPSC-Derived Hippocampal Spheroids: An Innovative Tool for
Stratifying Alzheimer Disease Patient-Specific Cellular Phenotypes and
Developing Therapies.” Stem Cell Reports, vol. 15, no. 1, pp. 256-273,
2020. View at: Publisher
Site | PubMed
[110]
Janise N Kuehner, Junyu Chen, Emily C Bruggeman, et
al. “5-Hydroxymethylcytosine Is Dynamically Regulated during Forebrain Organoid
Development and Aberrantly Altered in Alzheimer’s Disease.” Cell Rep,
vol. 35, no. 4, pp. 109042, 2021. View at: Publisher Site | PubMed
[111]
Cesar Gonzalez, Enrique Armijo, Javiera Bravo-Alegria,
et al. “Modeling Amyloid Beta and Tau Pathology in Human Cerebral Organoids.”
Mol Psychiatry, vol. 23, no. 12, pp. 2363-2374, 2018. View at: Publisher
Site | PubMed
[112]
Helen H Zhao, Gabriel G Haddad “Alzheimer’s Disease
like Neuropathology in Down Syndrome Cortical Organoids.” Front Cell
Neurosci, vol. 16, pp. 1050432, 2022. View at: Publisher
Site | PubMed
[113]
Yuan-Ta Lin, Jinsoo Seo, Fan Gao, et al. “APOE4 Causes
Widespread Molecular and Cellular Alterations Associated with Alzheimer’s
Disease Phenotypes in Human IPSC-Derived Brain Cell Types.” Neuron, vol.
98, no. 6, pp. 1141-1154, 2018. View at: Publisher Site | PubMed
[114]
Katharina Meyer, Heather M Feldman, Tao Lu, et al.
“REST and Neural Gene Network Dysregulation in IPSC Models of Alzheimer’s
Disease.” Cell Rep, vol. 26, no. 5, pp. 1112-1127, 2019. View at: Publisher
Site | PubMed
[115]
Shichao Huang, Zhen Zhang, Junwei Cao, et al.
“Chimeric Cerebral Organoids Reveal the Essentials of Neuronal and Astrocytic
APOE4 for Alzheimer’s Tau Pathology.” Signal Transduct Target Ther, vol.
7, no. 1, pp. 176, 2022. View at: Publisher Site | PubMed
[116]
Han-Kyu Lee, Clara Velazquez Sanchez, Mei Chen, et al.
“Three Dimensional Human Neuro-Spheroid Model of Alzheimer’s Disease Based on
Differentiated Induced Pluripotent Stem Cells.” PLoS One, vol. 11, no.
9, pp. e0163072, 2016. View at: Publisher Site | PubMed
[117]
Mei Chen, Han-Kyu Lee, Lauren Moo, et al. “Common
Proteomic Profiles of Induced Pluripotent Stem Cell-Derived Three-Dimensional
Neurons and Brain Tissue from Alzheimer Patients.” J Proteomics, vol.
182, pp. 21-33, 2018. View at: Publisher Site | PubMed
[118]
Jong-Chan Park, So-Yeong Jang, Dongjoon Lee, et al. “A
Logical Network-Based Drug-Screening Platform for Alzheimer’s Disease
Representing Pathological Features of Human Brain Organoids.” Nat Commun,
vol. 12, no. 1, pp. 280, 2021. View at: Publisher Site | PubMed
[119]
Ling Zhang, Shuli Sheng, Chuan Qin “The Role of HDAC6
in Alzheimer’s Disease.” J Alzheimers Dis, vol. 33, no. 2, pp. 283-295,
2013. View at: Publisher
Site | PubMed
[120]
Heesun Choi, Haeng Jun Kim, Jinhee Yang, et al.
“Acetylation Changes Tau Interactome to Degrade Tau in Alzheimer’s Disease
Animal and Organoid Models.” Aging Cell, vol. 19, no. 1, pp. e13081,
2020. View at: Publisher Site | PubMed
[121]
Damián Hernández, Louise A Rooney, Maciej Daniszewski,
et al. “Culture Variabilities of Human IPSC-Derived Cerebral Organoids Are a
Major Issue for the Modelling of Phenotypes Observed in Alzheimer’s Disease.” Stem
Cell Rev Rep, vol. 18, no. 2, pp. 718-731, 2022. View at: Publisher
Site | PubMed
[122]
Lalitha Venkataraman, Summer R Fair, Craig A McElroy,
et al. “Modeling Neurodegenerative Diseases with Cerebral Organoids and Other
Three-Dimensional Culture Systems: Focus on Alzheimer’s Disease.” Stem Cell
Rev Rep, vol. 18, no. 2, pp. 696-717, 2022. View at: Publisher
Site | PubMed
[123]
Jing Zhao, Wenyan Lu, Yingxue Ren, et al.
“Apolipoprotein E Regulates Lipid Metabolism and α-Synuclein Pathology in Human IPSC-Derived Cerebral Organoids.” Acta Neuropathol, vol. 142, no. 5, pp. 807-825, 2021. View at: Publisher
Site | PubMed
[124]
Erwan Lambert, Orthis Saha, Bruna Soares Landeira, et
al. “The Alzheimer Susceptibility Gene BIN1 Induces Isoform-Dependent
Neurotoxicity through Early Endosome Defects.” Acta Neuropathol Commun,
vol. 10, no. 1, pp. 4, 2022. View at: Publisher Site | PubMed
[125]
Dario Brunetti, Janniche Torsvik, Cristina Dallabona,
et al. “Defective PITRM 1 Mitochondrial Peptidase Is Associated with Aβ Amyloidotic Neurodegeneration.” EMBO Mol Med, vol. 8, no. 3, pp. 176-190, 2016. View at: Publisher
Site | PubMed
[126]
María José Pérez, Dina Ivanyuk, Vasiliki
Panagiotakopoulou, et al. “Loss of Function of the Mitochondrial Peptidase
PITRM1 Induces Proteotoxic Stress and Alzheimer’s Disease-like Pathology in
Human Cerebral Organoids.” Mol Psychiatry, vol. 26, no. 10, pp.
5733-5750, 2021. View at: Publisher Site | PubMed
[127]
Eric E Abrahamson, Wenxiao Zheng, Vaishali
Muralidaran, et al. “Modeling Aβ42
Accumulation in Response to Herpes Simplex Virus 1 Infection: 2D or 3D?” J Virol, vol. 95, no. 5, pp. e02219-e02220, 2021. View at: Publisher
Site | PubMed
[128]
Haowen Qiao, Wen Zhao, Moujian Guo, et al. “Cerebral
Organoids for Modeling of HSV-1-Induced-Amyloid β Associated Neuropathology and Phenotypic Rescue.” Int J Mol Sci, vol. 23, no. 11, pp. 5981, 2022. View at: Publisher
Site | PubMed
[129]
Ju-Hyun Lee, Geon Yoo, Juhyun Choi, et al. “Cell-Line
Dependency in Cerebral Organoid Induction: Cautionary Observations in Alzheimer’s
Disease Patient-Derived Induced Pluripotent Stem Cells.” Mol Brain, vol.
15, no. 1, pp. 46, 2022. View at: Publisher Site | PubMed
[130]
Samudyata, Ana O Oliveira, Susmita
Malwade, et al. “SARS-CoV-2 Promotes Microglial Synapse
Elimination in Human Brain Organoids.” Mol Psychiatry, vol. 27, no. 10,
pp. 3939-3950, 2022. View at: Publisher Site | PubMed
[131]
Alison Abbott “Are Infections Seeding Some Cases of
Alzheimer’s Disease.” Nature, vol. 587, no. 7832, pp. 22-25, 2020. View
at: Publisher
Site | PubMed
[132]
Serena Pavoni, Rafika Jarray, Ferid Nassor, et al.
“Small-Molecule Induction of Aβ-42 Peptide
Production in Human Cerebral Organoids to Model Alzheimer’s Disease Associated Phenotypes.” PLoS One, vol. 13, no. 12, pp.
e0209150, 2018. View at: Publisher
Site | PubMed
[133]
Jennifer Yejean Kim, Hyunkyung Mo, Juryun Kim, et al.
“Mitigating Effect of Estrogen in Alzheimer’s Disease-Mimicking Cerebral
Organoid.” Front Neurosci, vol. 16, pp. 816174, 2022. View at: Publisher
Site | PubMed
[134]
Ru Zhang, Juan Lu, Gang Pei, et al. “Galangin Rescues
Alzheimer’s Amyloid-β Induced
Mitophagy and Brain Organoid Growth Impairment.” Int J Mol
Sci, vol. 24, no. 4, pp. 3398, 2023. View at: Publisher
Site | PubMed
[135]
Xianwei Chen, Guoqiang Sun, E Tian, et al. “Modeling
Sporadic Alzheimer’s Disease in Human Brain Organoids under Serum Exposure.” Adv
Sci (Weinh), vol. 8, no. 18, pp. e2101462, 2021. View at: Publisher
Site | PubMed
[136]
Jinsoo Seo, Oleg Kritskiy, L Ashley Watson, et al.
“Inhibition of P25/Cdk5 Attenuates Tauopathy in Mouse and IPSC Models of
Frontotemporal Dementia.” J Neurosci, vol. 37, no. 41, pp. 9917-9924,
2017. View at: Publisher
Site | PubMed
[137]
Ferid Nassor, Rafika Jarray, Denis S F Biard, et al.
“Long Term Gene Expression in Human Induced Pluripotent Stem Cells and Cerebral
Organoids to Model a Neurodegenerative Disease.” Front Cell Neurosci,
vol. 14, pp. 14, 2020. View at: Publisher Site | PubMed
[138]
Kimberly L Fiock, Martin E Smalley, John F Crary, et
al. “Increased Tau Expression Correlates with Neuronal Maturation in the
Developing Human Cerebral Cortex.” eNeuro, vol. 7, no. 3, pp.
ENEURO.0058-20.2020, 2020. View at: Publisher Site | PubMed
[139]
Ivan Alić, Pollyanna A Goh, Aoife Murray, et al. “Patient-Specific
Alzheimer-like Pathology in Trisomy 21 Cerebral Organoids Reveals BACE2 as a
Gene Dose-Sensitive AD Suppressor in Human Brain.” Mol Psychiatry, vol.
26, no. 10, pp. 5766-5788, 2021. View at: Publisher Site | PubMed
[140]
Juan Luo, Hailin Zou, Yibo Guo, et al. “BACE2 Variant
Identified from HSCR Patient Causes AD-like Phenotypes in hPSC-Derived Brain
Organoids.” Cell Death Discov, vol. 8, no. 1, pp. 47, 2022. View at: Publisher
Site | PubMed
[141]
Hoi-Khoanh Giong, Manivannan Subramanian, Kweon Yu, et
al. “Non-Rodent Genetic Animal Models for Studying Tauopathy: Review of
Drosophila, Zebrafish and c. Elegans Models.” Int J Mol Sci, vol. 22,
no. 16, pp. 8465, 2021. View at: Publisher Site | PubMed
[142]
Elizabeth Di Lullo, Arnold R Kriegstein “The Use of
Brain Organoids to Investigate Neural Development and Disease.” Nat Rev
Neurosci, vol. 18, no. 10, pp. 573-584, 2017. View at: Publisher
Site | PubMed
[143]
Haihua Ma, Juan Chen, Zhiyu Deng, et al. “Multiscale
Analysis of Cellular Composition and Morphology in Intact Cerebral Organoids.” Biology
(Basel), vol. 11, no. 9, pp. 1270, 2022. View at: Publisher
Site | PubMed
[144]
Eugenio Cavalli, Giuseppe Battaglia,
Maria Sofia Basile, et al. “Exploratory Analysis of -Derived
Neuronal Cells as Predictors of Diagnosis and Treatment of Alzheimer Disease.” Brain
Sci, vol. 10, no. 3, pp. 166, 2020. View at: Publisher Site | PubMed
[145]
Sally Esmail, Wayne R Danter “NEUBOrg: Artificially
Induced Pluripotent Stem Cell-Derived Brain Organoid to Model and Study
Genetics of Alzheimer’s Disease Progression.” Front Aging Neurosci, vol.
13, pp. 643889, 2021. View at: Publisher Site | PubMed
[146]
Jung Yeon Lim, Jung Eun Lee, Soon A Park, et al.
“Protective Effect of Human-Neural-Crest-Derived Nasal Turbinate Stem Cells
against Amyloid-β
Neurotoxicity through Inhibition of Osteopontin in a Human Cerebral Organoid Model
of Alzheimer’s Disease.” Cells, vol. 11, no. 6, pp. 1029, 2022. View at:
Publisher
Site | PubMed
[147]
Gregory Jensen, Christian Morrill, Yu Huang “3D Tissue
Engineering, an Emerging Technique for Pharmaceutical Research.” Acta Pharm
Sin B, vol. 8, pp. 756-766, 2018. View at: Publisher Site | PubMed
[148]
Lu Qian, Julia Tcw “Human IPSC-Based Modeling of
Central Nerve System Disorders for Drug Discovery.” Int J Mol Sci, vol.
22, no. 3, pp. 1203, 2021. View at: Publisher Site | PubMed
[149]
Karl Grenier, Jennifer Kao, Phedias Diamandis
“Three-Dimensional Modeling of Human Neurodegeneration: Brain Organoids Coming
of Age.” Mol Psychiatry, vol. 25, no. 2, pp. 254-274, 2020. View at: Publisher
Site | PubMed
[150]
Nicolas Rouleau, William L Cantley, Volha
Liaudanskaya, et al. “A Long-Living Bioengineered Neural Tissue Platform to
Study Neurodegeneration.” Macromol Biosci, vol. 20, no. 3, pp. e2000004,
2020. View at: Publisher
Site | PubMed
[151]
Jonathan A Brassard, Matthias P Lutolf “Engineering
Stem Cell Self-Organization to Build Better Organoids.” Cell Stem Cell,
vol. 24, no. 6, pp. 860-876, 2019. View at: Publisher Site | PubMed
[152]
Sherida M de Leeuw, Stephanie Davaz, Debora Wanner, et
al. “Increased Maturation of IPSC-Derived Neurons in a Hydrogel-Based 3D
Culture.” J Neurosci Methods, vol. 360, pp. 109254, 2021. View at: Publisher
Site | PubMed
[153]
Yean Ju Hong, So Been Lee 1, Joonhyuk Choi, et al. “A
Simple Method for Generating Cerebral Organoids from Human Pluripotent Stem
Cells.” Int J Stem Cells, vol. 15, no. 1, pp. 95-103, 2022. View at: Publisher Site | PubMed
[154]
Selina Wray “Modelling Neurodegenerative Disease Using
Brain Organoids.” Semin Cell Dev Biol, vol. 111, pp. 60-66, 2021. View
at: Publisher
Site | PubMed
[155]
Xin Dong, Shi-Bo Xu, Xin Chen, et al. “Human Cerebral
Organoids Establish Subcortical Projections in the Mouse Brain after
Transplantation.” Mol Psychiatry, vol. 26, no. 7, pp. 2964-2976, 2021.
View at: Publisher
Site | PubMed
[156]
Yujin Ahn, Ju-Hyun An, Hae-Jun Yang, et al. “Human
Blood Vessel Organoids Penetrate Human Cerebral Organoids and Form a
Vessel-like System.” Cells, vol. 10, no. 8, pp. 2036, 2021. View at: Publisher
Site | PubMed
[157]
Bilal Cakir, Yangfei Xiang, Yoshiaki Tanaka, et al.
“Engineering of Human Brain Organoids with a Functional Vascular-like System.” Nat
Methods, vol. 16, no. 11, pp. 1169-1175, 2019. View at: Publisher
Site | PubMed
[158]
Onju Ham, Yeung Bae Jin, Janghwan Kim, et al. “Blood
Vessel Formation in Cerebral Organoids Formed from Human Embryonic Stem Cells.”
Biochem Biophys Res Commun, vol. 521, no. 1, pp. 84-90, 2020. View at: Publisher
Site | PubMed
[159]
Yingchao Shi, Le Sun, Mengdi Wang, et al.
“Vascularized Human Cortical Organoids (VOrganoids) Model Cortical Development in
vivo.” PLoS Biol, vol. 18, no. 5, 2020. View at: Publisher
Site | PubMed
[160]
Xin-Yao Sun, Xiang-Chun Ju, Yang Li, et al.
“Generation of Vascularized Brain Organoids to Study Neurovascular
Interactions.” Elife, vol. 11, pp. e76707, 2022. View at: Publisher
Site | PubMed
[161]
Chul Soon Park, Le Phuong Nguyen, Dongeun Yong
“Development of Colonic Organoids Containing Enteric Nerves or Blood Vessels
from Human Embryonic Stem Cells.” Cells, vol. 9, no. 10, pp. 2209, 2020.
View at: Publisher
Site | PubMed
[162]
Lotta E Oikari, Rucha Pandit, Romal Stewart, et al.
“Altered Brain Endothelial Cell Phenotype from a Familial Alzheimer Mutation
and Its Potential Implications for Amyloid Clearance and Drug Delivery.” Stem
Cell Reports, vol. 14, no. 5, pp. 924-939, 2020. View at: Publisher
Site | PubMed
[163]
JiSoo Park, Bo Kyeong Lee, Gi Seok Jeong, et al.
“Three-Dimensional Brain-on-a-Chip with an Interstitial Level of Flow and Its
Application as an in Vitro Model of Alzheimer’s Disease.” Lab Chip, vol.
15, no. 1, pp. 141-150, 2015. View at: Publisher Site | PubMed
[164]
Mohammed R Shaker, Julio Aguado, Harman Kaur Chaggar,
et al. “Klotho Inhibits Neuronal Senescence in Human Brain Organoids.” NPJ
Aging Mech Dis, vol. 7, no. 1, pp. 18, 2021. View at: Publisher
Site | PubMed
[165]
Adam A Sivitilli, Jessica T Gosio, Bibaswan Ghoshal,
et al. “Robust Production of Uniform Human Cerebral Organoids from Pluripotent
Stem Cells.” Life Sci Alliance, vol. 3, no. 5, pp. e202000707, 2020.
View at: Publisher
Site | PubMed
[166]
Sudha R Guttikonda, Lisa Sikkema, Jason Tchieu, et al.
“Fully Defined Human Pluripotent Stem Cell-Derived Microglia and Tri-Culture
System Model C3 Production in Alzheimer’s Disease.” Nat Neurosci, vol.
24, no. 3, pp. 343-354, 2021. View at: Publisher Site | PubMed
[167] Celia Luchena, Jone Zuazo-Ibarra, Jorge Valero, et al. “A Neuron, Microglia, and Astrocyte Triple Co-Culture Model to Study Alzheimer’s Disease.” Front Aging Neurosci, vol. 14, pp. 844534, 2022. View at: Publisher Site | PubMed
[168] Rebecca M Marton, Yuki Miura, Steven A Sloan, et al. “Differentiation and Maturation of Oligodendrocytes in Human Three-Dimensional Neural Cultures.” Nat Neurosci, vol. 22, no. 3, pp. 484-491, 2019. View at: Publisher Site | PubMed
